July 20, 2021
Strabismus, a common disorder of ocular alignment, affects 3% to 5% of children in the United States, yet the etiology of childhood strabismus is poorly understood. Risk factors for ocular misalignment among a minority of children include premature birth, maternal tobacco exposure, and developmental or neurological disorders. How prominent the role of genetics might be in the development of the most prevalent forms of childhood strabismus, however, is currently unknown.
To investigate candidate genes associated with familial strabismus and propose a theory of their interaction in familial strabismus associated with early neurodevelopment, a research team including Brian G. Mohney, M.D., a consultant in Ophthalmology at Mayo Clinic in Rochester, Minnesota, used exome sequencing analysis to identify possible risk variants of familial strabismus.
"Recent advances in next-generation sequencing technologies have allowed the identification of disease-causing genes," says Dr. Mohney. "Whole-exome sequencing, or WES, is a well-established technology for identifying variants within the coding regions, or exons, of known genes. WES provides an opportunity to identify causal genes in Mendelian and complex genetic disorders with high sensitivity and specificity. Familial studies using WES have revealed rare variants associated with strabismus or genetic syndromes with strabismus." In their study, published in Genes (Basel) in 2021, the research team used WES to detect single nucleotide variants (SNVs) and insertions-deletions (indels) within a cohort of families with two or more members affected with comitant strabismus.
Researchers prospectively recruited children with familial strabismus, defined as having two or more family members with horizontal, comitant strabismus. The proband and available family members were examined for the following:
- Visual acuity
- Angle of deviation at distance (3 meters) and at near (1/3 meter) measured with the prism and alternate cover test
- Stereopsis
- Cycloplegic refraction
Affected members with strabismus were defined as having a horizontal, comitant misalignment of 10 or more prism diopters at either distance or near in the primary position. Nonaffected individuals were defined as having orthophoria and normal stereopsis.
Among the 98 participants initially recruited, 87 participants remained for downstream analysis: Researchers evaluated 18 families — comprising 53 patients diagnosed with strabismus and 34 unaffected family members — using WES, with a 53.6X average sequence read depth and 88.5% average call rate.
Data set results included a total of 320,470 unique SNVs (approximately 120,299 SNVs per individual) and 51,072 unique indels (approximately 18,607 indels per individual). Of these results, 84,376 SNVs and 3,050 indels were located in protein-coding sequences, while 236,094 SNVs and 48,022 indels were found in noncoding sequences.
To identify candidate genes for causing strabismus, the researchers defined causal variants
by the following criteria:
- Occurring rarely in the general population
- Resulting in either a loss of function (including nonsense, frameshift and canonical splicing site change) or missense
- Missense variants predicted to be deleterious by either the scale-invariant feature transform or the PolyPhen2 algorithm
- Observed in genes with the probability of being loss of function intolerant (score 0.9 or greater)
- Characterized in the developmental process
Pedigree information with risk variants
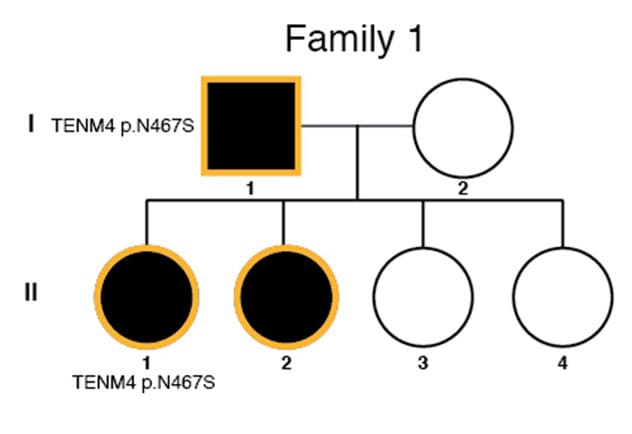
Pedigree information with risk variants
To assess credible risk variants associated with familial strabismus, researchers prioritized risk variants segregated in multiple family members or occurring in multiple families, possibly suggesting their role in familial strabismus. "From 18 families with horizontal and comitant strabismus, we identified 60 candidate variants, including three loss-of-function and 57 missense variants in 58 genes. Two genes had multiple variants," says Dr. Mohney.
Prioritization of the most credible risk variants showed clear segregation in family members affected by strabismus. "We found risk variants in four genes — FAT3, KCNH2, CELSR1 and TTYH1 — in five families, suggesting their role in development of familial strabismus," says Dr. Mohney.
Several families in the study had different clinical phenotypes even though they had the same genetic variation, supporting the possibility that the type of strabismus can be affected not only by the role of genetic variation in the expression of strabismus but also by the developmental process, the time of ocular misalignment occurrence or environmental factors.
"Next-generation genome sequencing approaches hold great promise in identifying rare variants associated with the disorder and elucidating the genetic causes of strabismus," says Dr. Mohney. "Despite our small-sample study, we were able to identify four genes with causative associations with strabismus.
"We also observed genetic heterogeneity in strabismus cases with risk genes occurring in several neurodevelopmental disorders. These results add to the increasing evidence for genetic heterogeneity within familial comitant strabismus."
For more information
An JY, et al. Identification of possible risk variants of familial strabismus using exome sequencing analysis. Genes (Basel). 2021;12:75.